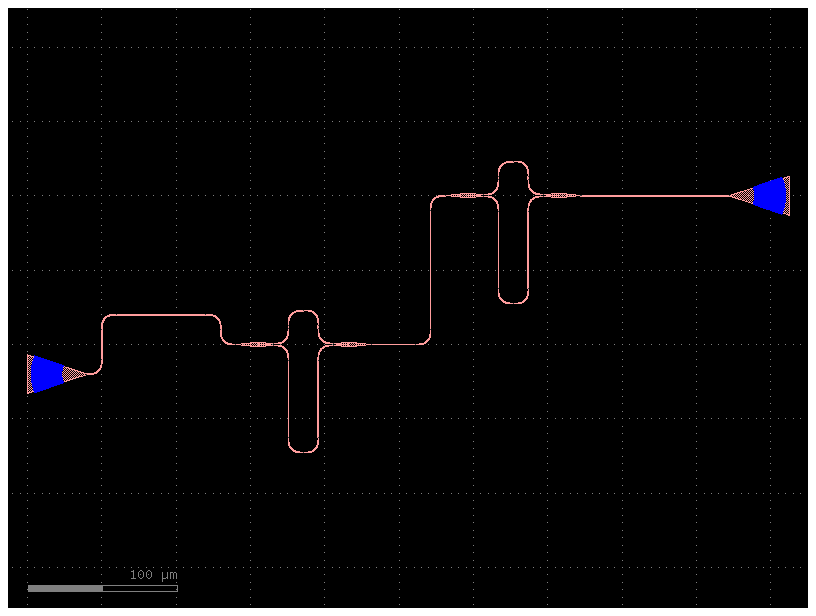
You have two options for working with gdsfactory:
- python flow: you define your layout using python functions (Parametric Cells), and connect them with routing functions.
- YAML Place and AutoRoute: you define your Component as Place and Route in YAML. From the netlist you can simulate the Component or generate the layout.
YAML is a human readable version of JSON that you can use to define placements and routes
to define a a YAML Component you need to define:
- instances: with each instance setting
- placements: with X and Y
And optionally:
- routes: between instance ports
- connections: to connect instance ports to other ports (without routes)
- ports: define input and output ports for the top level Component.
gdsfactory VSCode extension has a filewatcher for *.pic.yml files that will show them live in klayout as you edit them.
The extension provides you with useful code snippets and filewatcher extension to see live modifications of *pic.yml or *.py files. Look for the telescope button on the top right of VSCode 🔭.
import gdsfactory as gf
from IPython.display import Code
filepath = "yaml_pics/pads.pic.yml"
Code(filepath, language="yaml+jinja")gf.read.from_yaml(filepath).plot()2023-12-09 17:32:14.327 | WARNING | gdsfactory.pdk:get_active_pdk:721 - No active PDK. Activating generic PDK.
2023-12-09 17:32:14.627 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
2023-12-09 17:32:14.628 | INFO | gdsfactory.pdk:activate:334 - 'generic' PDK is now active
2023-12-09 17:32:14.721 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
/home/runner/work/gdsfactory/gdsfactory/gdsfactory/component.py:1584: UserWarning: Unnamed cells, 1 in 'Unnamed_774052fb$1'
gdspath = component.write_gds(logging=False)
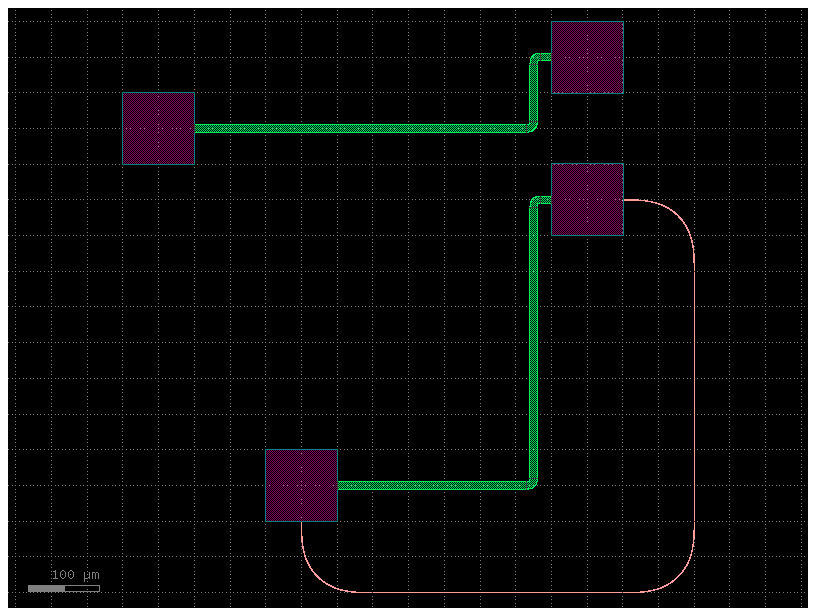
Lets start by defining the instances and placements section in YAML
Lets place an mmi_long where you can place the o1 port at x=20, y=10
filepath = "yaml_pics/mmis.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:15.519 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
/home/runner/work/gdsfactory/gdsfactory/gdsfactory/component.py:1584: UserWarning: Unnamed cells, 1 in 'Unnamed_f4f1eca5$1'
gdspath = component.write_gds(logging=False)
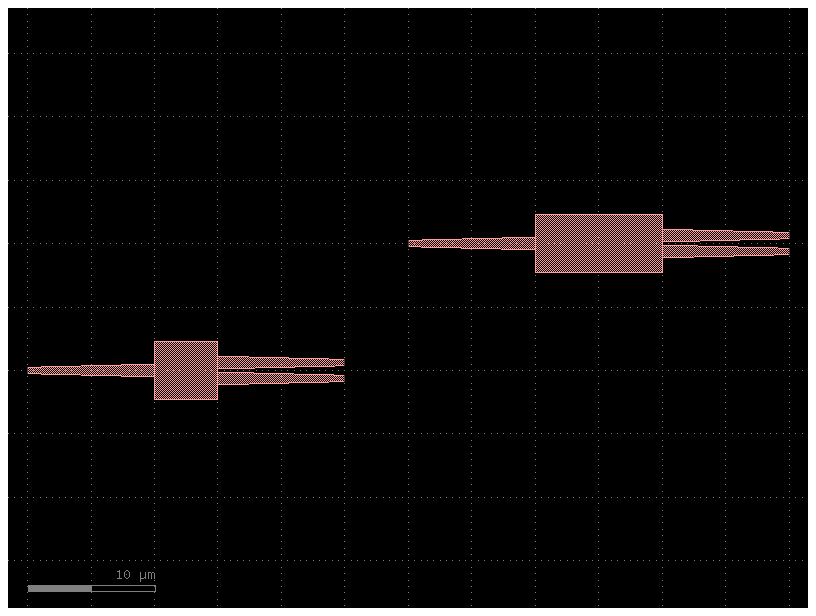
ports¶
You can expose any ports of any instance to the new Component with a ports section in YAML
Lets expose all the ports from mmi_long into the new component.
Ports are exposed as new_port_name: instance_name, port_name
filepath = "yaml_pics/ports_demo.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:15.832 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
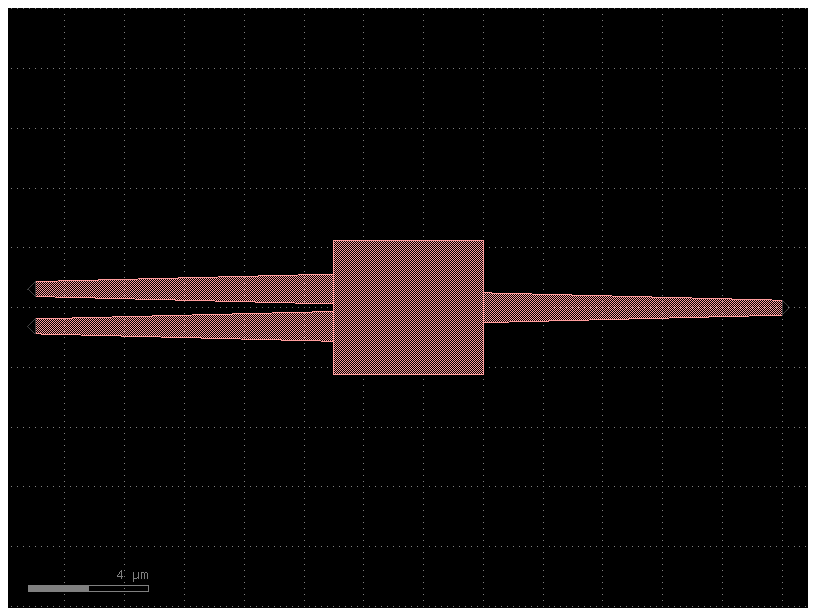
You can also define a mirror placement using a port
Try mirroring with other ports o2, o3 or with a number as well as with a rotation 90, 180, 270
filepath = "yaml_pics/mirror_demo.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:16.123 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
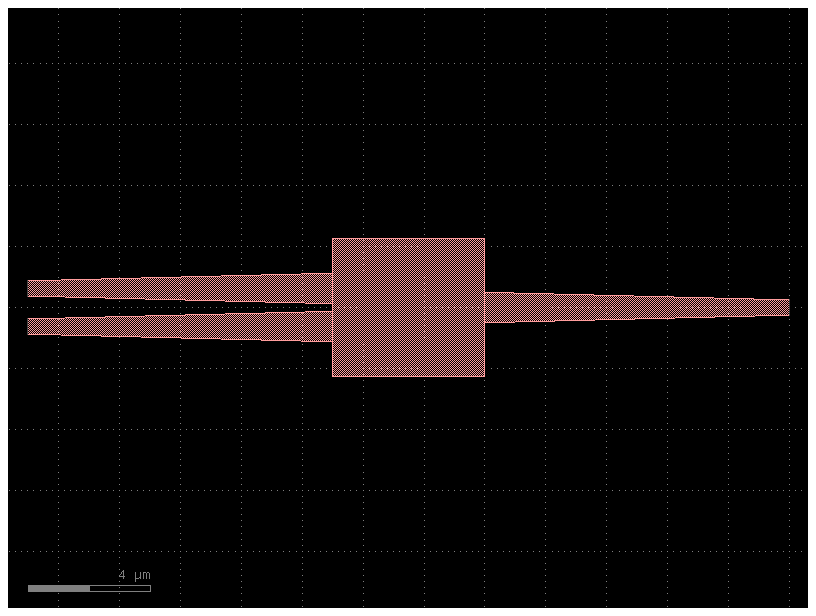
connections¶
You can connect any two instances by defining a connections section in the YAML file.
it follows the syntax instance_source,port : instance_destination,port
filepath = "yaml_pics/connections_demo.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:16.411 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
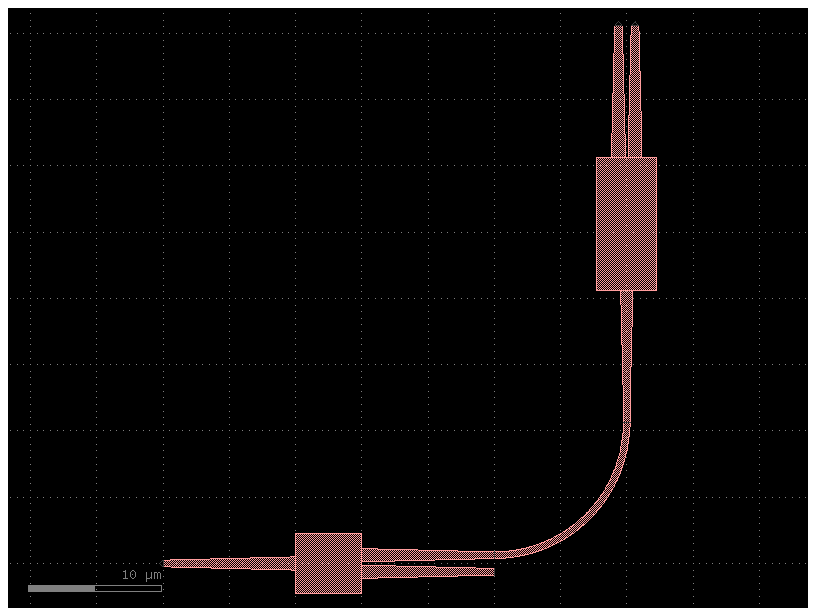
Relative port placing
You can also place a component with respect to another instance port
You can also define an x and y offset with dx and dy
filepath = "yaml_pics/relative_port_placing.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:16.714 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
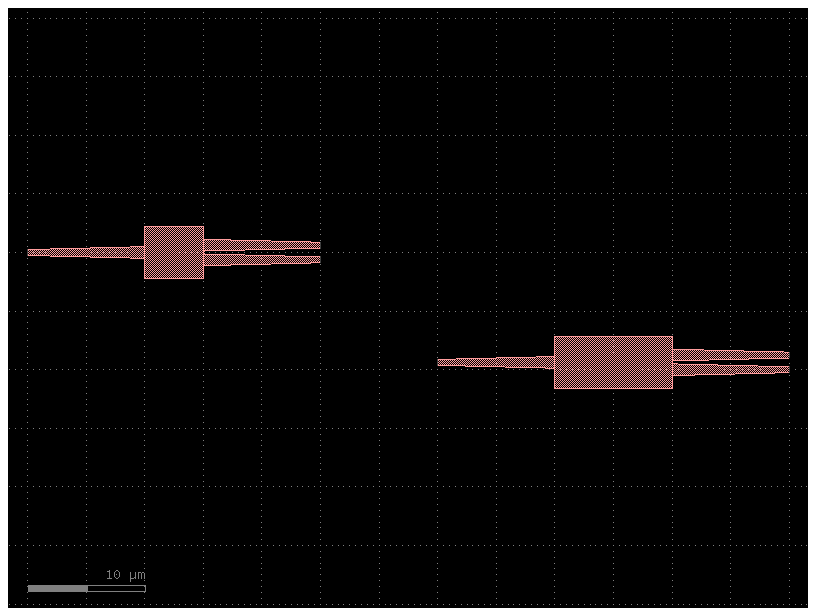
routes¶
You can define routes between two instances by defining a routes section in YAML
it follows the syntax
routes:
route_name:
links:
instance_source,port: instance_destination,port
settings: # for the route (optional)
waveguide: strip
width: 1.2
filepath = "yaml_pics/routes.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:17.106 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
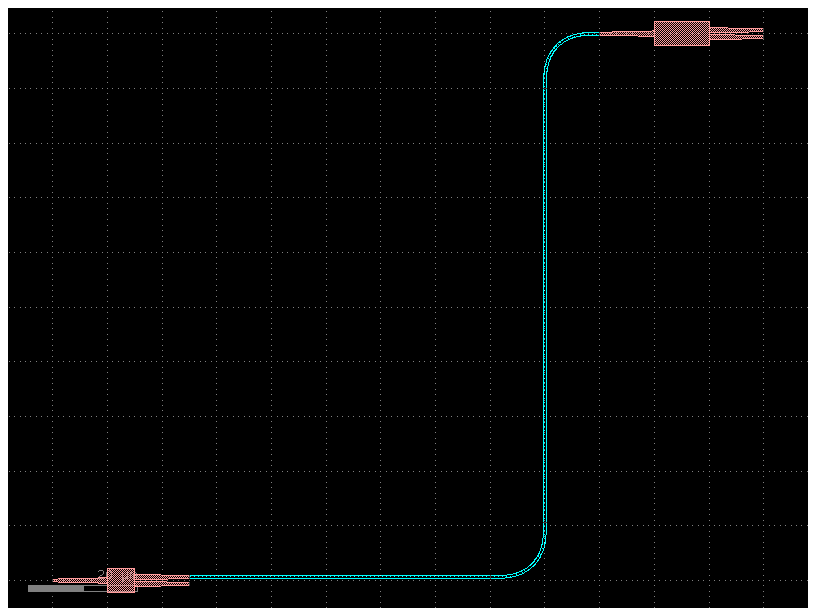
instances, placements, connections, ports, routes¶
Lets combine all you learned so far.
You can define the netlist connections of a component by a netlist in YAML format
Note that you define the connections as instance_source.port -> instance_destination.port so the order is important and therefore you can only
change the position of the instance_destination
You can define several routes that will be connected using gf.routing.get_bundle
filepath = "yaml_pics/routes_mmi.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:17.443 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
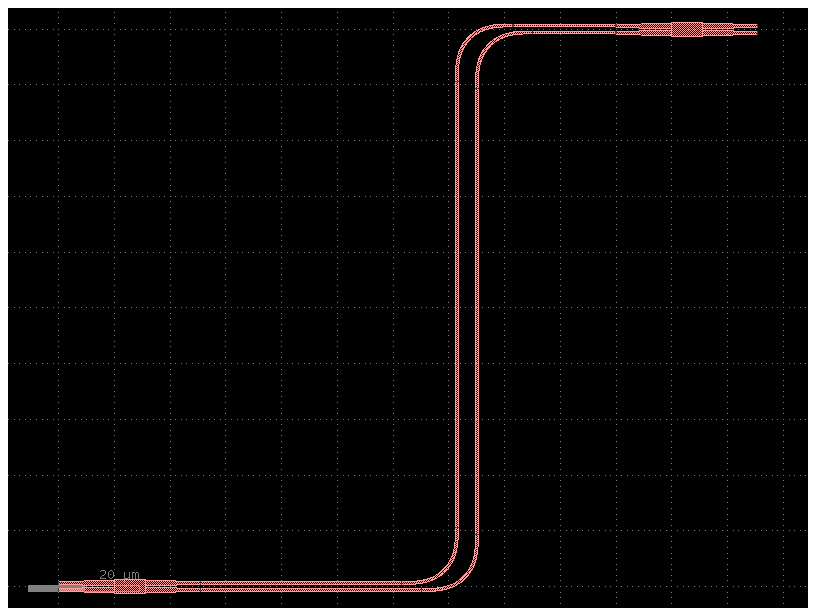
You can also add custom component_factories to gf.read.from_yaml
@gf.cell
def pad_new(size=(100, 100), layer=(1, 0)):
c = gf.Component()
compass = c << gf.components.compass(size=size, layer=layer)
c.ports = compass.ports
return c
gf.get_active_pdk().register_cells(pad_new=pad_new)
c = pad_new()
c.plot()
filepath = "yaml_pics/new_factories.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:18.009 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
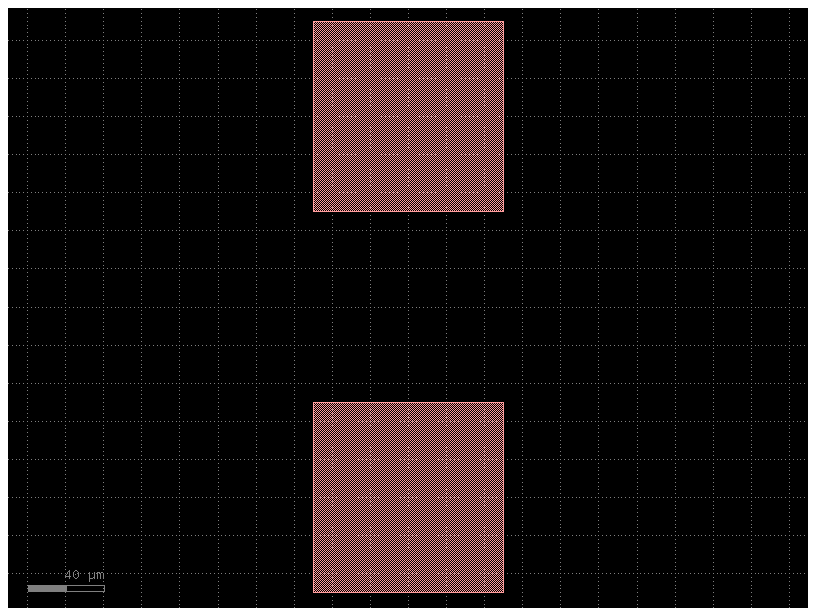
filepath = "yaml_pics/routes_custom.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:18.306 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
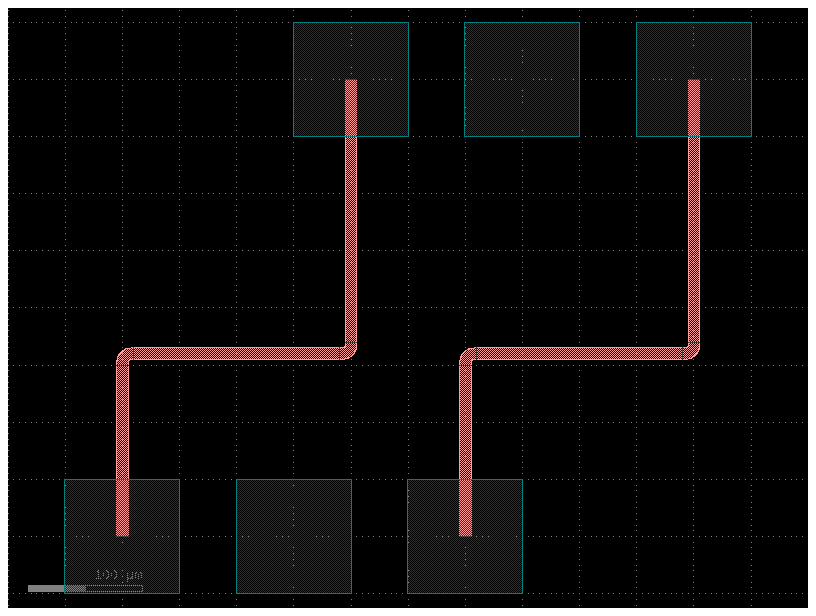
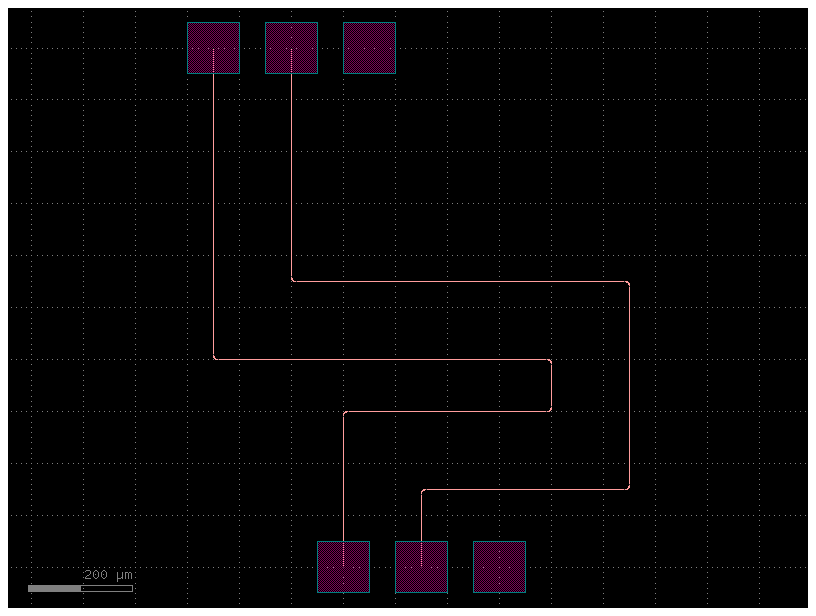
Also, you can define route bundles with different settings and specify the route factory as a parameter as well as the settings for that particular route alias.
filepath = "yaml_pics/pads_path_length_match.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:18.654 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
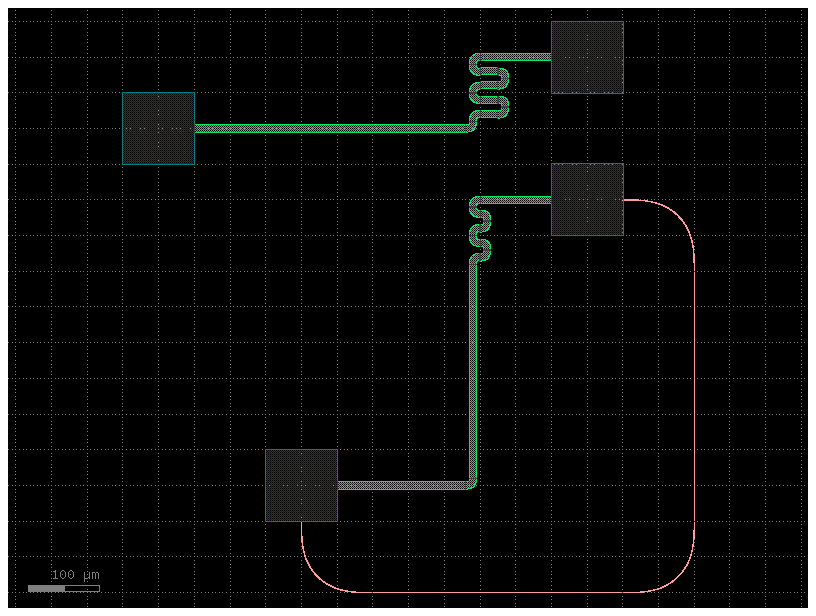
filepath = "yaml_pics/routes_path_length_match.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:19.036 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
/home/runner/work/gdsfactory/gdsfactory/gdsfactory/component.py:1584: UserWarning: Unnamed cells, 1 in 'Unnamed_0315283a$1'
gdspath = component.write_gds(logging=False)
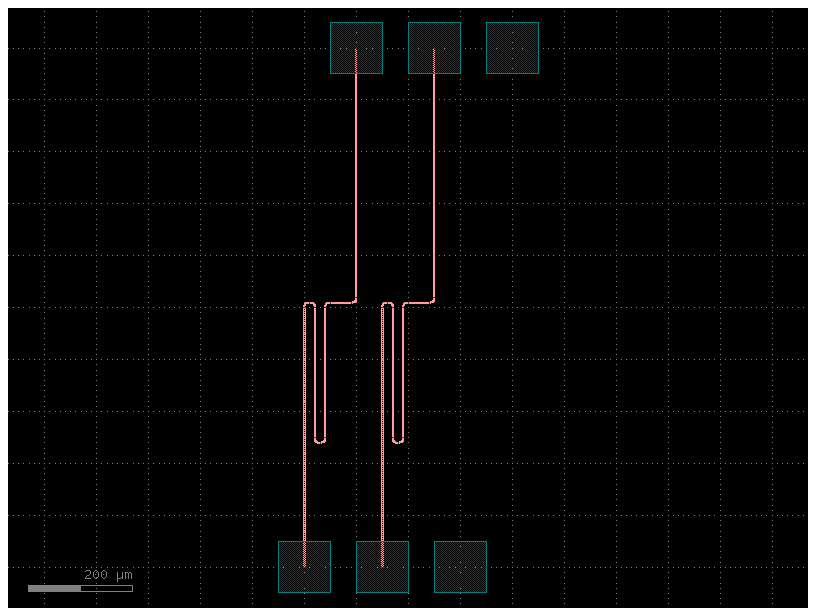
filepath = "yaml_pics/routes_waypoints.pic.yml"
Code(filepath, language="yaml+jinja")c = gf.read.from_yaml(filepath)
c.plot()2023-12-09 17:32:19.510 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
/home/runner/work/gdsfactory/gdsfactory/gdsfactory/component.py:1584: UserWarning: Unnamed cells, 1 in 'Unnamed_c057659c$1'
gdspath = component.write_gds(logging=False)
Jinja Pcells¶
You use jinja templates in YAML cells to define Pcells.
from IPython.display import Code
from gdsfactory.read import cell_from_yaml_template
gf.clear_cache()
jinja_yaml = """
default_settings:
length_mmi:
value: 10
description: "The length of the long MMI"
width_mmi:
value: 5
description: "The width of both MMIs"
instances:
mmi_long:
component: mmi1x2
settings:
width_mmi: {{ width_mmi }}
length_mmi: {{ length_mmi }}
mmi_short:
component: mmi1x2
settings:
width_mmi: {{ width_mmi }}
length_mmi: 5
connections:
mmi_long,o2: mmi_short,o1
ports:
o1: mmi_long,o1
o2: mmi_short,o2
o3: mmi_short,o3
"""
pic_filename = "demo_jinja.pic.yml"
with open(pic_filename, mode="w") as f:
f.write(jinja_yaml)
pic_cell = cell_from_yaml_template(pic_filename, name="demo_jinja")
gf.get_active_pdk().register_cells(
demo_jinja=pic_cell
) # let's register this cell so we can use it later
Code(filename=pic_filename, language="yaml+jinja")You’ll see that this generated a python function, with a real signature, default arguments, docstring and all!
help(pic_cell)Help on function demo_jinja:
demo_jinja(*, length_mmi=10, width_mmi=5)
demo_jinja: a templated yaml cell. This cell accepts keyword arguments only
Keyword Args:
length_mmi: The length of the long MMI
width_mmi: The width of both MMIs
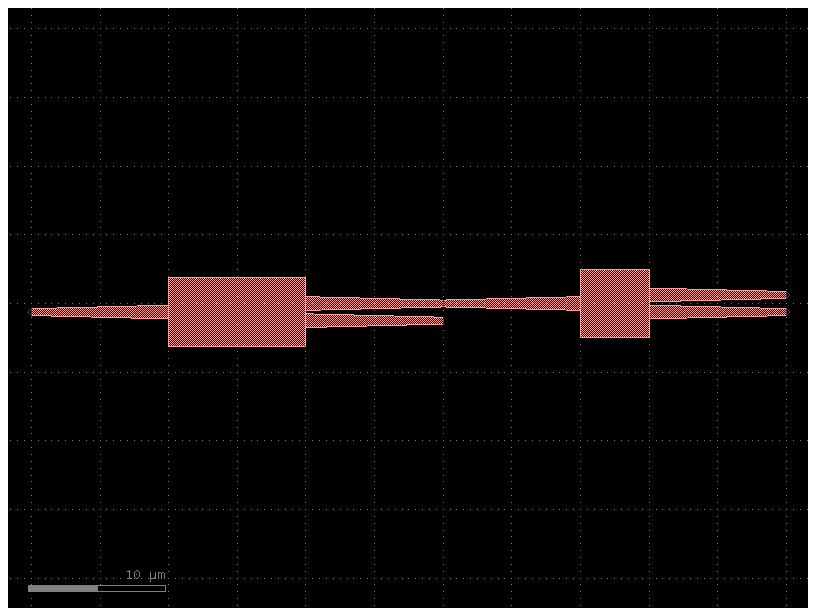
You can invoke this cell without arguments to see the default implementation
c = pic_cell()
c.plot()2023-12-09 17:32:19.893 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
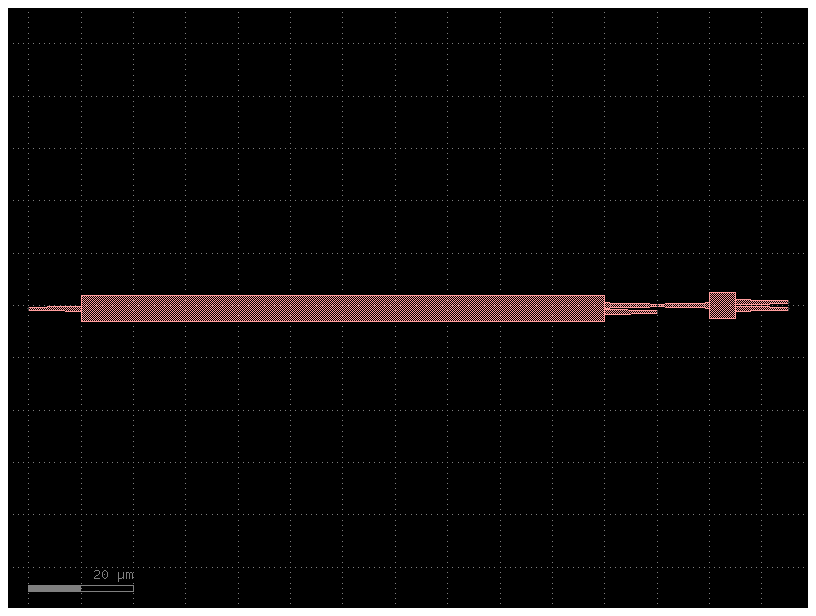
Or you can provide arguments explicitly, like a normal cell. Note however that yaml-based cells only accept keyword arguments, since yaml dictionaries are inherently unordered.
c = pic_cell(length_mmi=100)
c.plot()2023-12-09 17:32:20.172 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
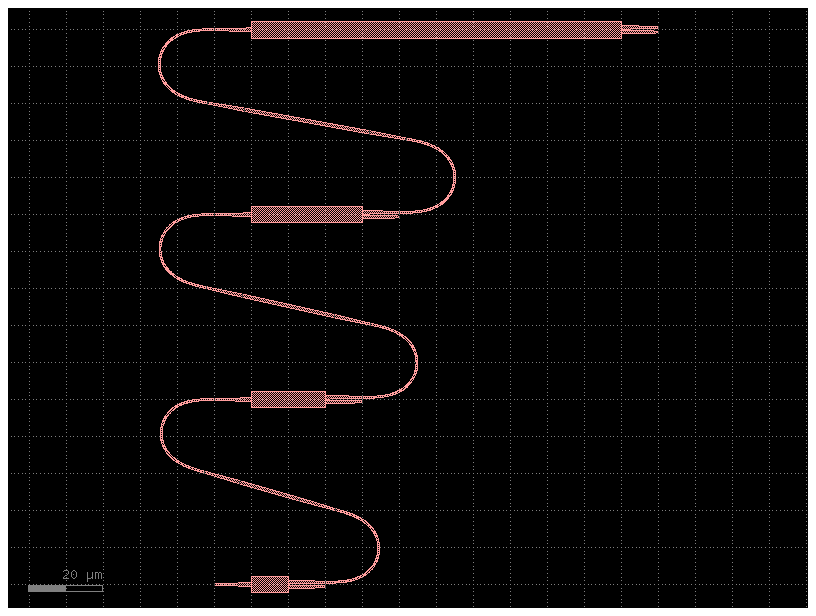
The power of jinja-templated cells become more apparent with more complex cells, like the following.
gf.clear_cache()
jinja_yaml = """
default_settings:
length_mmis:
value: [10, 20, 30, 100]
description: "An array of mmi lengths for the DOE"
spacing_mmi:
value: 50
description: "The vertical spacing between adjacent MMIs"
mmi_component:
value: mmi1x2
description: "The mmi component to use"
instances:
{% for i in range(length_mmis|length)%}
mmi_{{ i }}:
component: {{ mmi_component }}
settings:
width_mmi: 4.5
length_mmi: {{ length_mmis[i] }}
{% endfor %}
placements:
{% for i in range(1, length_mmis|length)%}
mmi_{{ i }}:
port: o1
x: mmi_0,o1
y: mmi_0,o1
dy: {{ spacing_mmi * i }}
{% endfor %}
routes:
{% for i in range(1, length_mmis|length)%}
r{{ i }}:
routing_strategy: get_bundle_all_angle
links:
mmi_{{ i-1 }},o2: mmi_{{ i }},o1
{% endfor %}
ports:
{% for i in range(length_mmis|length)%}
o{{ i }}: mmi_{{ i }},o3
{% endfor %}
"""
pic_filename = "demo_jinja_loops.pic.yml"
with open(pic_filename, mode="w") as f:
f.write(jinja_yaml)
big_cell = cell_from_yaml_template(pic_filename, name="demo_jinja_loops")
Code(filename=pic_filename, language="yaml+jinja")bc = big_cell()
bc.plot()2023-12-09 17:32:20.489 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
bc2 = big_cell(
length_mmis=[10, 20, 40, 100, 200, 150, 10, 40],
spacing_mmi=60,
mmi_component="demo_jinja",
)
bc2.plot()2023-12-09 17:32:21.249 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
2023-12-09 17:32:21.343 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
2023-12-09 17:32:21.449 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
2023-12-09 17:32:21.546 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
2023-12-09 17:32:21.762 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
2023-12-09 17:32:21.861 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
2023-12-09 17:32:21.968 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.

In general, the jinja-yaml parser has a superset of the functionalities and syntax of the standard yaml parser. The one notable exception is with settings. When reading any yaml files with settings blocks, the default settings will be read and applied, but they will not be settable, as the jinja parser has a different mechanism for setting injection with the default_settings block and jinja2.
filepath = "yaml_pics/mzi_lattice_filter.pic.yml"
mzi_lattice = cell_from_yaml_template(filepath, name="mzi_lattice_filter")
Code(filepath, language="yaml")c = mzi_lattice(delta_length=10)
c.plot()2023-12-09 17:32:23.114 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.
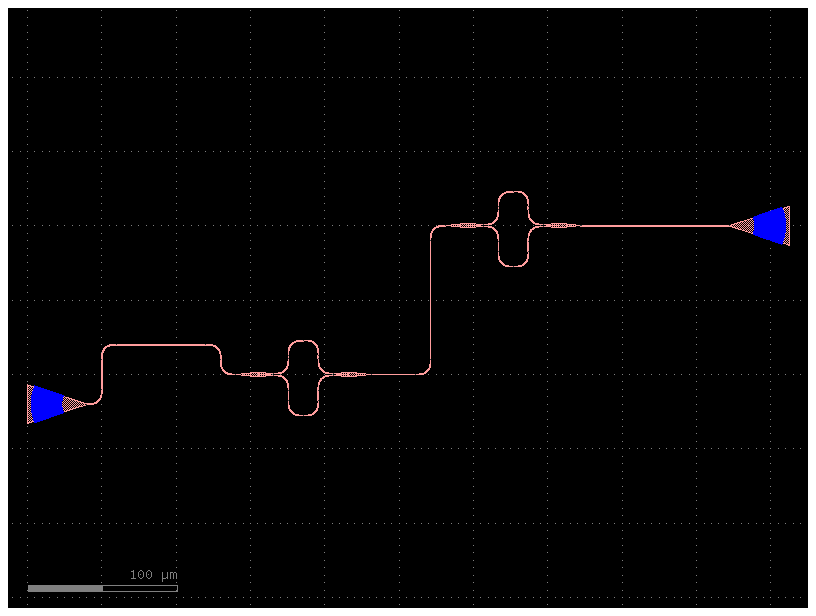
c = mzi_lattice(delta_length=100)
c.plot()2023-12-09 17:32:23.572 | INFO | gdsfactory.technology.layer_views:__init__:790 - Importing LayerViews from YAML file: '/home/runner/work/gdsfactory/gdsfactory/gdsfactory/generic_tech/layer_views.yaml'.